SeekOne DD scFAST-seq Kit
SeekOne® Digital Droplet (SeekOne® DD) High-Throughput scFAST-seq Kit
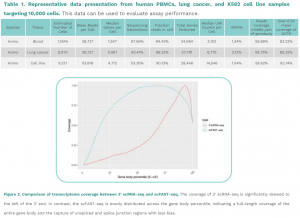
Application: Ideal for microRNA and viral RNA profiling from single-cell libraries. It can process up to 12,000 cells, creating full-length RNA libraries for single-cell RNA sequencing (scRNA-seq).
Key Features:
SeekOne® Digital Droplet (SeekOne® DD) High-Throughput scFAST-seq Kit Comprehensive RNA Profiling: Captures both polyadenylated and non-polyadenylated transcripts.
- Full-Length Libraries: Ideal for studying alternative splicing and non-coding RNAs.
- Enhanced Transcript Detection: Robustly identifies viral RNAs and miRNAs from lncRNAs.
- Multi-Omics Integration: Combines mutation detection and gene expression for tumor heterogeneity analysis.

RNA Types Detected:
- lncRNA, circRNA, miRNA, and viral RNA.
Captures unique miRNAs like miR-21, miR-155, and miR-122, linked to cancer, cardiovascular, and neurological diseases.
Detects non-polyadenylated viral RNAs, providing insights into influenza, rabies, HDV, and other viruses.
Applications in Disease Research:
Cancer: Profiling miRNAs (e.g., miR-21, miR-34a) for tumor growth and suppression.
Cardiovascular Diseases: Detection of miRNAs related to heart function (e.g., miR-1, miR-133).
Neurological Disorders: Analysis of miRNAs (e.g., miR-132, miR-124) linked to brain development and neurodegenerative diseases.
Viral Research: Identifies RNA structures of viruses, including influenza and Ebola, aiding in the study of virus-host interactions.
SeekOne® DD scFAST-seq: An all-in-one solution integrating various RNA analysis methods, enabling high-resolution single-cell profiling for oncology, virology, and transcriptomics research.
Author:
Lieven Gevaert, Gentaur Laboratories, Leuven, Belgium